Multi-party Record Linkage with Blocking¶
In this tutorial, we will demonstrate the CLI tools for multiparty record linkage with blocking techniques.
[1]:
import io
import os
import math
import time
from IPython import display
import json
from collections import defaultdict
import pandas as pd
import anonlink
import clkhash
from clkhash import clk
from blocklib import assess_blocks_2party
from anonlinkclient.utils import combine_clks_blocks, deserialize_bitarray, deserialize_filters
%run util.py
Suppose we are interested to find records that appear at least twice in 3 parties
Generate CLKs and Candidate Blocks¶
First we have a look at dataset
[2]:
# NBVAL_IGNORE_OUTPUT
corruption_rate = 20
file_template = 'data/ncvr_numrec_5000_modrec_2_ocp_' + str(corruption_rate) + '_myp_{}_nump_10.csv'
df1 = pd.read_csv(file_template.format(0))
df1.head()
[2]:
| recid | givenname | surname | suburb | postcode | |
|---|---|---|---|---|---|
| 0 | 1503359 | pauline | camkbell | lilescille | 28091 |
| 1 | 1972058 | deborah | galyen | ennike | 286z3 |
| 2 | 889525 | charle5 | mitrhell | roaring river | 28669 |
| 3 | 4371845 | petehr | werts | swannanoa | 28478 |
| 4 | 1187991 | katpy | silbiger | duyham | 27705 |
A linkage schema instructs clkhash how to treat each column for generating CLKs. A detailed description of the linkage schema can be found in the api docs. We will ignore the column ‘recid’ for CLK generation.
[3]:
# NBVAL_IGNORE_OUTPUT
with open("novt_schema.json") as f:
print(f.read())
{
"version": 3,
"clkConfig": {
"l": 1024,
"kdf": {
"type": "HKDF",
"hash": "SHA256",
"salt": "SCbL2zHNnmsckfzchsNkZY9XoHk96P/G5nUBrM7ybymlEFsMV6PAeDZCNp3rfNUPCtLDMOGQHG4pCQpfhiHCyA==",
"info": "c2NoZW1hX2V4YW1wbGU=",
"keySize": 64
}
},
"features": [
{
"identifier": "recid",
"ignored": true
},
{
"identifier": "givenname",
"format": {
"type": "string",
"encoding": "utf-8",
"maxLength": 30,
"case": "lower"
},
"hashing": {
"comparison": {"type": "ngram", "n": 2},
"strategy": {"bitsPerFeature": 100},
"hash": {"type": "blakeHash"},
"missingValue": {
"sentinel": ".",
"replaceWith": ""
}
}
},
{
"identifier": "surname",
"format": {
"type": "string",
"encoding": "utf-8",
"maxLength": 30,
"case": "lower"
},
"hashing": {
"comparison": {"type": "ngram", "n": 2},
"strategy": {"bitsPerFeature": 100},
"hash": {"type": "blakeHash"},
"missingValue": {
"sentinel": ".",
"replaceWith": ""
}
}
},
{
"identifier": "suburb",
"format": {
"type": "string",
"encoding": "utf-8",
"maxLength": 30,
"case": "lower"
},
"hashing": {
"comparison": {"type": "ngram", "n": 2},
"strategy": {"bitsPerFeature": 100},
"hash": {"type": "blakeHash"},
"missingValue": {
"sentinel": ".",
"replaceWith": ""
}
}
},
{
"identifier": "postcode",
"format": {
"type": "string",
"encoding": "utf-8",
"maxLength": 30,
"case": "lower"
},
"hashing": {
"comparison": {"type": "ngram", "n": 2},
"strategy": {"bitsPerFeature": 100},
"hash": {"type": "blakeHash"},
"missingValue": {
"sentinel": ".",
"replaceWith": ""
}
}
}
]
}
Validate the schema¶
The command line tool can check that the linkage schema is valid:
[4]:
# NBVAL_IGNORE_OUTPUT
!anonlink validate-schema "novt_schema.json"
schema is valid
Hash data¶
We can now hash our Personally Identifiable Information (PII) data from the CSV file using our defined linkage schema. We must provide a secret key to this command - this key has to be used by both parties hashing data. For this toy example we will use the secret ‘secret’, for real data, make sure that the secret contains enough entropy, as knowledge of this secret is sufficient to reconstruct the PII information from a CLK!
[5]:
secret = 'secret'
[6]:
# NBVAL_IGNORE_OUTPUT
!anonlink hash 'data/ncvr_numrec_5000_modrec_2_ocp_20_myp_0_nump_10.csv' secret 'novt_schema.json' 'novt_clk_0.json'
CLK data written to novt_clk_0.json
Let’s hash data for party B and C:
[7]:
# NBVAL_IGNORE_OUTPUT
!anonlink hash 'data/ncvr_numrec_5000_modrec_2_ocp_20_myp_1_nump_10.csv' secret 'novt_schema.json' 'novt_clk_1.json'
CLK data written to novt_clk_1.json
[8]:
# NBVAL_IGNORE_OUTPUT
!anonlink hash 'data/ncvr_numrec_5000_modrec_2_ocp_20_myp_2_nump_10.csv' secret 'novt_schema.json' 'novt_clk_2.json'
CLK data written to novt_clk_2.json
anonlink provides a command describe to inspect the hashing results i.e. the Cryptographic Longterm Key (CLK). Normally we will expect a relative symmetric shape popcount with a moderate mean comparing to bloom filter length.
[9]:
# NBVAL_IGNORE_OUTPUT
!anonlink describe 'novt_clk_0.json'
----------------------------------------------------------------------------------------------------------------------------
| popcounts |
----------------------------------------------------------------------------------------------------------------------------
298| o
282| o ooooo
267| o ooooo
251| ooooooo
235| ooooooooo o
220| ooooooooo o
204| ooooooooo o
188| o ooooooooo oo
173| ooooooooooo oo o
157| ooooooooooo oooo
141| oooooooooooo ooooo
126| oooooooooooo ooooo
110| ooooooooooooo ooooo
94| oooooooooooooo ooooo
79| ooooooooooooooo ooooooo
63| oooooooooooooooo ooooooo
47| oooooooooooooooooo ooooooo o
32| o ooooooooooooooooooo oooooooooo
16| oooo ooooooooooooooooooo oooooooooooo
1| o o o oooooooooooo ooooooooooooooooooo oooooooooooooooo o
-------------------------------------------------------------
2 2 2 2 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3
9 9 9 9 0 0 0 0 0 1 1 1 1 1 2 2 2 2 2 3 3 3 3 3 3 4 4 4 4 4 5
4 5 7 9 1 3 5 7 9 1 3 4 6 8 0 2 4 6 8 0 2 3 5 7 9 1 3 5 7 9 1
. . . . . . . . . . . . . . . . . . . . . . . . . . .
9 8 7 6 5 4 3 2 1 9 8 7 6 5 4 3 2 1 9 8 7 6 5 4 3 2 1
------------------------
| Summary |
------------------------
| observations: 5000 |
|min value: 294.000000 |
| mean : 328.055000 |
|max value: 351.000000 |
------------------------
In this case, the popcount mean is not very large compared to bloom filter length (1024). If the popcount mean is large, you can reduce it by modifying the schema. For more details, please have a look at this tutorial.
Block dataset¶
Blocking is a technique that makes record linkage scalable. It is achieved by partitioning datasets into groups, called blocks and only comparing records in corresponding blocks. This can reduce the number of comparisons that need to be conducted to find which pairs of records should be linked.
Similar to the hashing above, the blocking is configured with a schema. For this linkage we chose ‘lambda-fold’ as blocking technique. This blocking method is proposed in paper An LSH-Based Blocking Approach with a Homomorphic Matching Technique for Privacy-Preserving Record Linkage. We also provide a detailed explanation of how this blocking method works in this tutorial.
[10]:
# NBVAL_IGNORE_OUTPUT
with open("blocking_schema.json") as f:
print(f.read())
{
"type": "lambda-fold",
"version": 1,
"config": {
"blocking-features": [1, 2],
"Lambda": 30,
"bf-len": 2048,
"num-hash-funcs": 5,
"K": 20,
"input-clks": true,
"random_state": 0
}
}
Party A Blocks its Data
[11]:
# NBVAL_IGNORE_OUTPUT
!anonlink block 'novt_clk_0.json' 'blocking_schema.json' 'novt_blocks_0.json'
Number of Blocks: 127903
Minimum Block Size: 1
Maximum Block Size: 33
Average Block Size: 1
Median Block Size: 1
Standard Deviation of Block Size: 0.655395485238156
Party B Blocks its Data
[12]:
# NBVAL_IGNORE_OUTPUT
!anonlink block 'novt_clk_1.json' 'blocking_schema.json' 'novt_blocks_1.json'
Number of Blocks: 127632
Minimum Block Size: 1
Maximum Block Size: 24
Average Block Size: 1
Median Block Size: 1
Standard Deviation of Block Size: 0.6600347712901997
Party C Blocks its Data
[13]:
# NBVAL_IGNORE_OUTPUT
!anonlink block 'novt_clk_2.json' 'blocking_schema.json' 'novt_blocks_2.json'
Number of Blocks: 127569
Minimum Block Size: 1
Maximum Block Size: 23
Average Block Size: 1
Median Block Size: 1
Standard Deviation of Block Size: 0.6707693358673655
Get Ground Truth¶
[14]:
#NBVAL_IGNORE_OUTPUT
truth = []
for party in [0, 1, 2]:
df = pd.read_csv('data/ncvr_numrec_5000_modrec_2_ocp_20_myp_{}_nump_10.csv'.format(party))
truth.append(pd.DataFrame({'id{}'.format(party): df.index, 'recid': df['recid']}))
dfj = truth[0].merge(truth[1], on='recid', how='outer')
for df in truth[2:]:
dfj = dfj.merge(df, on='recid', how='outer')
dfj = dfj.drop(columns=['recid'])
true_matches = set()
for row in dfj.itertuples(index=False):
cand = [(i, int(x)) for i, x in enumerate(row) if not math.isnan(x)]
if len(cand) > 1:
true_matches.add(tuple(cand))
print(f'we have {len(true_matches)} true matches')
e = iter(true_matches)
for i in range(10):
print(next(e))
we have 1649 true matches
((0, 159), (1, 169), (2, 169))
((1, 2309), (2, 2137))
((0, 366), (1, 589), (2, 362))
((0, 3719), (1, 3701), (2, 1560))
((0, 182), (1, 758), (2, 760))
((0, 3886), (1, 3865), (2, 311))
((0, 2878), (2, 280))
((0, 498), (1, 2269), (2, 492))
((0, 3630), (2, 282))
((1, 692), (2, 374))
Solve with Anonlink¶
[16]:
# NBVAL_IGNORE_OUTPUT
clk_files = ['novt_clk_{}.json'.format(x) for x in range(3)]
block_files = ['novt_blocks_{}.json'.format(x) for x in range(3)]
clk_blocks = []
for i, (clk_f, block_f) in enumerate(zip(clk_files, block_files)):
print('Combining CLKs and Blocks for Party {}'.format(i))
clk_blocks.append(json.load(combine_clks_blocks(open(clk_f, 'rb'), open(block_f, 'rb')))['clknblocks'])
clk_groups = []
rec_to_blocks = {}
for i, clk_blk in enumerate(clk_blocks):
clk_groups.append(deserialize_filters([r[0] for r in clk_blk]))
rec_to_blocks[i] = {rind: clk_blk[rind][1:] for rind in range(len(clk_blk))}
Combining CLKs and Blocks for Party 0
Combining CLKs and Blocks for Party 1
Combining CLKs and Blocks for Party 2
Assess Linkage Quality¶
We can assess the linkage quality by precision and recall.
- Precision is measured by the proportion of found record groups classified as true matches.
- Recall is measured by the proportion of true matching groups that are classified as found groups
[17]:
#NBVAL_IGNORE_OUTPUT
threshold = 0.87
# matching with blocking
found_groups = solve(clk_groups, rec_to_blocks, threshold)
print("Example found groups: ")
for i in range(10):
print(found_groups[i])
precision, recall = evaluate(found_groups, true_matches)
print('\n\nWith blocking: ')
print(f'precision: {precision}, recall: {recall}')
# matching without blocking
found_groups = naive_solve(clk_groups, threshold)
precision, recall = evaluate(found_groups, true_matches)
print('Without blocking: ')
print(f'precision: {precision}, recall: {recall}')
Example found groups:
((0, 755), (1, 1395), (2, 1392))
((0, 2492), (1, 3757), (2, 43))
((0, 3460), (1, 3657), (2, 266))
((1, 3227), (2, 454), (0, 136))
((0, 1066), (1, 1980), (2, 2003))
((1, 2572), (2, 4597))
((1, 2221), (2, 2516))
((0, 3219), (1, 2870))
((1, 758), (2, 760), (0, 182))
((0, 2194), (1, 2270), (2, 127))
With blocking:
precision: 0.7802197802197802, recall: 0.7319587628865979
Without blocking:
precision: 0.7808661926308985, recall: 0.7325651910248635
Assess Blocking¶
Reduction Ratio
Reduction ratio measures the proportion of number of comparisons reduced by using blocking technique. If we have two data providers each has 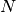 number of records, then
number of records, then
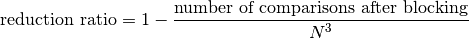
Set Completeness
Set completeness (aka pair completeness in two-party senario) measure how many true matches are maintained after blocking. It is evalauted as
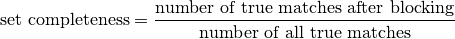
[18]:
# NBVAL_IGNORE_OUTPUT
block_a = json.load(open('novt_blocks_0.json'))['blocks']
block_b = json.load(open('novt_blocks_1.json'))['blocks']
block_c = json.load(open('novt_blocks_2.json'))['blocks']
filtered_reverse_indices = [block_a, block_b, block_c]
# filtered_reverse_indices[0]
data = []
for party in [0, 1, 2]:
dfa = pd.read_csv('data/ncvr_numrec_5000_modrec_2_ocp_0_myp_{}_nump_10.csv'.format(party))
recid = dfa['recid'].values
data.append(recid)
rr, reduced_num_comparison, naive_num_comparison = reduction_ratio(filtered_reverse_indices, data, K=2)
print('\nWith blocking, we reduced {:,} comparisons to {:,} comparisons i.e. the reduction ratio={}'
.format(naive_num_comparison, reduced_num_comparison, rr))
With blocking, we reduced 125,000,000,000 comparisons to 441,381 comparisons i.e. the reduction ratio=0.999996468952
[19]:
# NBVAL_IGNORE_OUTPUT
sc = set_completeness(filtered_reverse_indices, true_matches, K=2)
print('Set completeness = {}'.format(sc))
Set completeness = 0.9757428744693754